| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |34 |35 |36 |37 |38 |39 |40 |41 |42 |43 |44 |45 |review |
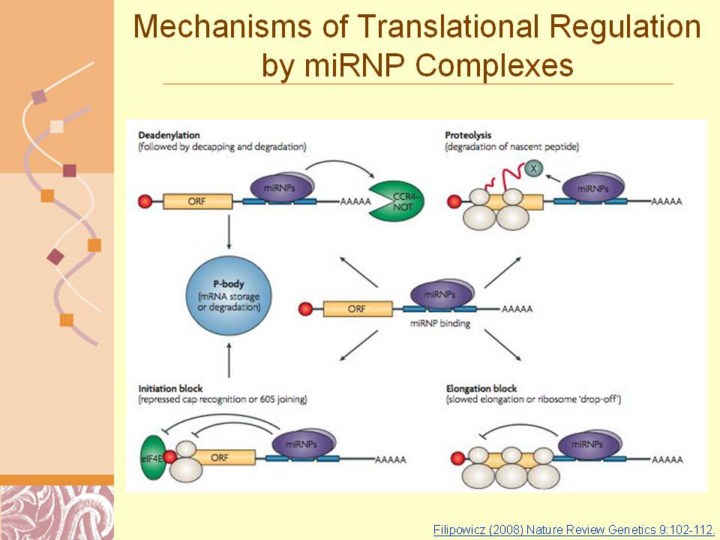 |
Filipowicz (2008) Nature Review Genetics 9:102-112.
Nature Reviews | Genetics Figure 3 | Possible mechanisms of the microRnA-mediated post-transcriptional gene repression in animal cells. Binding of micro-ribonucleoproteins (miRNPs), possibly complexed with accessory factors, to mRNA 3′ UTR can induce deadenylation and decay of target mRNAs56,78,79,83 (upper left). Alternatively, miRNPs can repress translation initiation at either the cap-recognition stage43,44,53–55 or the 60S subunit joining stage57 (bottom left). mRNAs repressed by deadenylation or at the translation-initiation stage are moved to P-bodies for either degradation or storage. The repression can also occur at post-initiation phases of translation66–68, owing to either slowed elongation or ribosome ‘drop-off’ (bottom right). Proteolytic cleavage of nascent polypeptides was also proposed as a mechanism of the miRNA-induced repression of protein production67 (upper-right). A protease (X) that might be involved in the process has not been identified. The 7-methylguanosine cap is represented by a red circle. eIF4E, eukaryotic initiation factor 4E. |