| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |34 |35 |36 |37 |38 |39 |40 |41 |42 |43 |44 |45 |review |
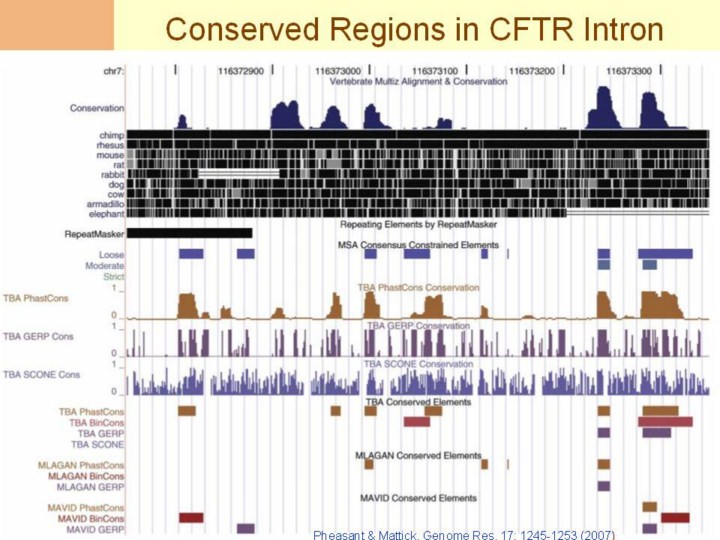 |
Pheasant & Mattick, Genome Res. 17: 1245-1253 (2007)
Conservation in the ENCODE CFTR locus. The diagram shows a 600-bp region in an intron of the ST7 gene (hg17 chr7:116372751– 116373350). The top panel (“Vertebrate Multiz Alignment & Conservation”) shows phastCons conservation scores based on 17-way alignments (Siepel et al. 2005). In black below this are alignments of human with chimp, rhesus, mouse, rat, rabbit, dog, cow, armadillo, and elephant. “Repeating Elements by RepeatMasker” shows an ancient repeat annotated as a MIR, which is 27% divergent from the MIR consensus, near the limit of detection. “MSA Consensus Constrained Elements” shows eight regions predicted to be conserved by at least one algorithm (“Loose” set), two regions predicted to be conserved by at least two algorithms in at least two alignments (“Moderate” set), and no regions predicted to be conserved by all algorithms in all alignments (“Strict” set). “TBA phastCons Conservation,” “TBA GERP Conservation,” and “TBA SCONE Conservation” show conservation scores over the TBA alignment from phastCons, GERP, and SCONE algorithms, respectively. “TBA Conserved Elements,” “MLAGAN Conserved Elements,” and “MAVID Conserved Elements” show elements predicted conserved based on the scores from the phastCons, BinCons, GERP, and SCONE algorithms across alignments from TBA, MLAGAN, and MAVID, respectively (Margulies et al. 2007) (image from http://genome.ucsc.edu/). The figure illustrates several difficulties in identifying selective constraints from regions that are not highly conserved: (1) conserved blocks are predicted within ARs assumed to evolve neutrally; (2) conservation scores vary depending on the species aligned (phastCons scores in the top panel are different from scores in TBA phastCons scores); (3) patterns of identified conservation vary between algorithms over the same alignment (compare the pattern of TBA scores from phastCons, GERP, and SCONE); and (4) conserved element predictions based on these scores vary between different algorithms on the same alignment as well as between the same algorithm over different alignments (compare phastCons, BinCons, and GERP elements over TBA, MLAGAN, and MAVID alignments). |