| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |34 |35 |36 |37 |38 |39 |40 |41 |42 |43 |44 |45 |review |
 |
Filipowicz (2008) Nature Review Genetics 9:102-112.
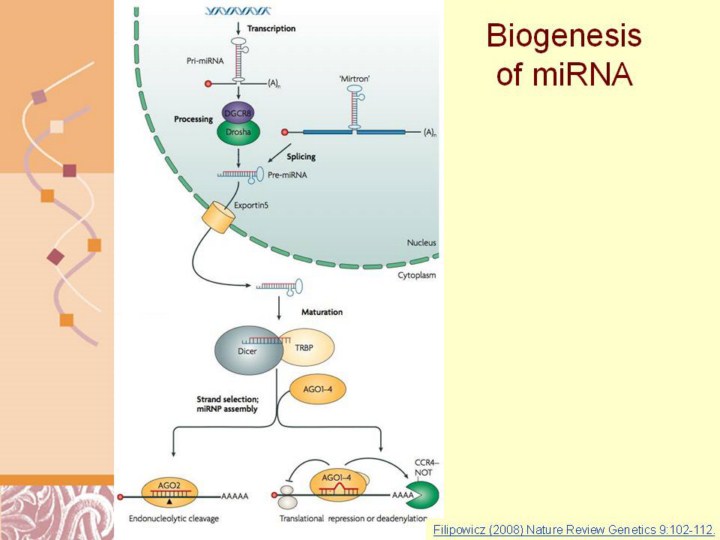
microRNAs (miRNAs) are processed from precursor molecules (pri- miRNAs), which are either transcribed from independent miRNA genes or are portions of introns of protein-coding RNA polymerase II transcripts. A single pri-miRNA often contains sequences for several different miRNAs. Pri-miRNAs fold into hairpin structures containing imperfectly base-paired stems and are processed in two steps, catalysed by the RNase III type endonucleases Drosha (also known as RN3) and Dicer. Both Drosha and Dicer function in complexes with proteins containing dsRNA-binding domains (dsRBDs). The Drosha partners are the pasha protein in Drosophila melanogaster or DiGeorge syndrome critical region gene 8 (DGCR8) in mammals. The Drosha–DGCR8 complex processes pri-miRNAs to ~70-nucleotide hairpins known as pre-miRNAs1,3,21,24. Some spliced-out introns in Caenorhabditis elegans, D. melanogaster and mammals correspond precisely to pre-miRNAs (mirtrons), thus circumventing the requirement for Drosha–DGCR8 (Refs 125–127). Plant genomes do not encode Drosha homologues, and all miRNA biogenesis steps in Arabidopsis thaliana are carried out by one of four Dicer-like proteins29. In animals, pre- miRNAs are transported to the cytoplasm by exportin5, where they are cleaved by Dicer (complexed with TAR RNA binding protein (TRBP) in mammals and the loquacious gene product in D. melanogaster) to yield ~20-bp miRNA duplexes. One strand is then selected to function as a mature miRNA, while the other strand is degraded. Occasionally, both arms of the pre-miRNA hairpin give rise to mature miRNAs1,3,21,24. Vertebrates and C. elegans contain single dicer genes, but some other organisms like D. melanogaster and plants express two or more Dicer proteins that function as heterodimers with different dsRBD proteins and have specialized functions1,3,21,24. Following their processing, miRNAs are assembled into ribonucleoprotein (RNP) complexes called micro-RNPs (miRNPs) or miRNA-induced silencing complexes (miRISCs). The assembly is a dynamic process, usually coupled with pre-miRNA processing by Dicer, but its details are not well understood1,3,21,24. The key components of miRNPs are proteins of the Argonaute (AGO) family. Of the many paralogues encoded in plant and metazoan genomes, usually only some — known as AGO proteins — function in miRNA or both miRNA and small interfering RNA (siRNA) pathways. In mammals, four AGO proteins (AGO1 to AGO4) function in the miRNA repression but only AGO2 functions in RNAi. In C. elegans, which expresses 27 Argonaute proteins, RDE1 is involved in RNAi and ALG1 and ALG2 function in the miRNA pathway24,25. Apart from AGOs, miRNPs can contain further proteins that function as regulatory factors or effectors mediating inhibitory function of miRNPs19,23,24. Examples are the fragile X mental retardation protein, FMRP, and its D. melanogaster orthologue, dFXR, which are RNA-binding proteins known to act as modulators of translation, particularly in neurons (reviewed in Ref. 128). Some P-body components such as GW182 and RCK/p54 (see BOX 4) interact with miRNP AGO proteins and are essential for inducing repression78,92,104. |