| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |34 |35 |36 |37 |38 |39 |40 |41 |42 |43 |44 |45 |review |
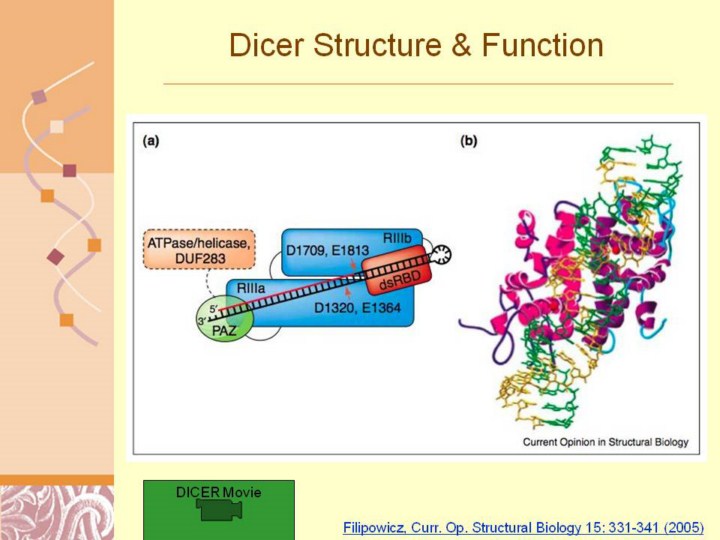 |
Filipowicz, Curr. Op. Structural Biology 15: 331-341 (2005)
Model of Dicer function. (a) Scheme summarizing a possible mechanism of pre-miRNA processing by human Dicer. The helicase/ATPase and DUF283 domains, with no assigned function, are delineated with a broken line. The enzyme contains a single dsRNA cleavage center with two independent catalytic sites. The center is formed by the RIIIa and RIIIb domains of the same Dicer molecule, and processes the dsRNA 20 bp from its terminus. The PAZ domain recognizes the 30 overhang end created by Drosha processing. The placement of the RIII domains illustrates the asymmetry of the catalytic region, with RIIIa cleaving the descending arm and RIIIb the ascending arm of the pre-miRNA hairpin [17]. (b) Model of bacterial (Aquifex aeolicus) RNase III interaction with dsRNA. The model is based on a crystal structure of the catalytic domain dimer [18] and the docking of the 30-bp dsRNA A helix, permitting cleavage of each RNA strand at phosphodiesters separated by 2 bp [17]. Two RNase III monomers are in different shades of purple. Mn2+ ions present in each catalytic center are shown as red spheres. The interaction of dsRNA with the intramolecular pseudo-dimer of the Dicer RIIIa and RIIIb domains is very probably similar. |