| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |34 |35 |36 |37 |38 |39 |40 |41 |42 |43 |44 |45 |46 |47 |48 |review |
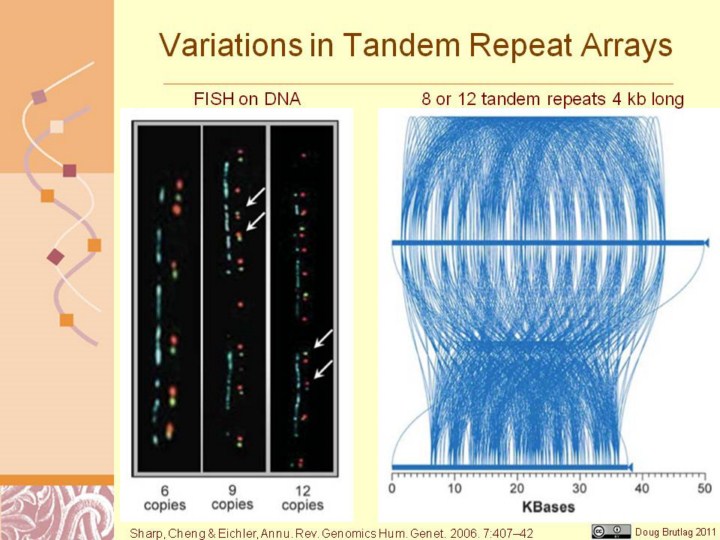 |
Visualization of tandem repeat structures in the human genome. (a) Fiber fluorescent in situ hybridization (FISH) demonstrates variable numbers of tandem repeats of the AMY1A gene on chromosome 1. High-resolution fiber FISH was performed on stretched DNA fibers using the BAC RP11–259N12 co-hybridized with a 5′ amylase gene probe (green) and a 3′ amylase gene probe (red ), revealing variable numbers of AMY1A tandem gene copies. Six, nine, and twelve gene signals were observed on different chromosomes. Note that the orientation of some gene copies appears inverted within the tandem array (arrowed), suggesting complexity induced by multiple rearrangements. The approximate length of the polymorphic region was estimated to vary from 150 kb to 425 kb in these three individuals (adapted from Reference 57). Reproduced with permission from Nature Publishing Group. (b) Miropeats (99) comparison of the sequence of fosmid WIBR2–1701E24 (16) with the corresponding region of hg17 of the human genome reference assembly (chr10:124,327,000–124,377,000). Lines connect regions of identity both within and between the human genome assembly (top) and the fosmid sequence (bottom), allowing the repeat structure to be visualized. This region contains a tandem motif with a repeat size of ∼4 kb, which shows variation in copy number from ∼8 to ∼12 between the two individuals sequenced.
|