| front |1 |2 |3 |4 |5 |6 |7 |8 |9 |10 |11 |12 |13 |14 |15 |16 |17 |18 |19 |20 |21 |22 |23 |24 |25 |26 |27 |28 |29 |30 |31 |32 |33 |review |
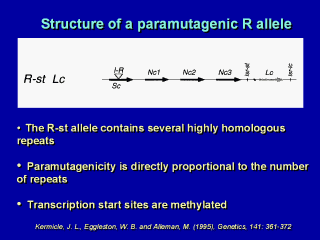 |
The structure of a paramutagenic allele of the
R locus is shown here. The key element is that this is a highly redundant locus that contains multiple copies of virtually identical coding sequences derived by unequal crossing-over. Kermicle and his colleagues identified 80 derivaties of this locus that arose by unequal crossingover and correlated the structure of the allele with its paramutagenicity -- a prodigious amount of work. These investigators found that the paramutagenicity of the derivatives was -- almost without exception -- proportional to the number of repeats. It had long been hypothesized that the transposon in the paramutagenic allele was responsible for its paramutagenicity, but derivatives lacking the transposon remain paramutagenic. |