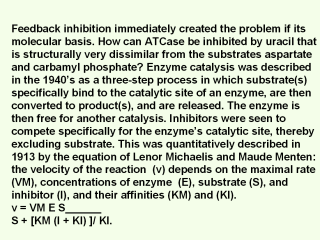
 |
Also, in
comparison to these classical asymptotic enzyme kinetics of rate vs.
substrate concentration, kinetics are cooperatively S-shaped,
for the isoleucine synthetic enzyme14 and for ATCase.15 This
suggests that these enzymes are composed of
interacting subunits. A pure enzyme was required to
understand these discrepancies, to eliminate
uncertainties posed by effects and explanations based on
complexities of properties and metabolism in crude extracts.
For starting material, ATCase was de-repressed 1,000
fold, and then purified and crystallized by Margaret
Shepherdson. With this pure enzyme the feedback
inhibitor was demonstrated to be not uridine but
cytidine triphosphate (CTP). But this compound has no apparent
structural similarity to the pathway’s initial substrates.16
This dilemma was solved by the discovery of enzyme’s
regulatory sites distinct from catalytic sites. An
initial demonstration of a regulatory site was that
ATP (which is not a substrate) increases ATPase
activity, in contrast to inhibitory CTP.15 This rules out the mechanism
of an interaction at the catalytic site, since binding
of ATP to the active site would have to be inhibitory.
It must therefore bind to a different, regulatory site.
Such binding of a regulatory molecule can change the
protein’s structure and thereby its catalytic site’s activity,
per the induced fit model proposed for enzymes by Daniel
Koshland.17 |
